Simple and fast characterization tool for biosensor development
UV-spectroscopy with Thermal Melt Analysis System – microvolumes in spectral focus
One of the key aspects in the development of DNA-based biosensors relies on understanding the molecular binding event to be detected. DNA is a polymer comprised of nucleotides forming four bases (adenine, thymine, guanine and cytosine). These nucleotides are bonded together via phosphodiester bonds. In a double helix, two strands of DNA are held together via hydrogen bonding between adenine and thymine with two bonds and guanine and cytosine with triple bonds respectively. The study of DNA is central to the understanding of many biological processes that have been identified in many human diseases.
At the University of Bath, the Biosensor Research Laboratory has been pioneering the development of DNA-based electrochemical sensors for the detection of a wide range of biomarkers ranging from DNA, microRNAs, proteins and cells. The addition of the UV-Vis spectrophotometer together with Thermal Melt Analysis System has been a very useful tool to understand the biosensor development approach.
Biosensor, signal and target DNA
A biosensor is an analytical device that can be used for detection of an analyte (pollutant, viral DNA, antigen or protein) by combining a biological component (enzyme, DNA, antibody or aptamer respectively) that acts like a probe and binds to the target. Such a binding event gives rise to a signal which is detected by a transducer (mass sensitive, optical or electrochemical) [1].
In order to get a signal, it is important that the specific molecular event occurs, which in the case of a DNA biosensor means the capture of the target DNA. Therefore, it becomes inevitable to have the right probe that binds specifically to the target. A controlled fabrication strategy is to monitor each step of the development of the biosensor. Hence, it is important to know if the probe which in this case is a single-stranded DNA and the target sequence are binding. Shimadzu UV-1800 with Thermal Melt Analysis System finds its place in such a development phase of the biosensor (figure 1).
 Figure 1: Shimadzu UV-1800 with Thermal Melt Analysis System (TMSPC-8) consisting of eight series micro multi-cell cuvette
Figure 1: Shimadzu UV-1800 with Thermal Melt Analysis System (TMSPC-8) consisting of eight series micro multi-cell cuvette
Nucleic acids, as single or double strands of DNA and RNA, absorb ultraviolet (UV) light due to the heterocyclic rings of the nucleotides. The absorption properties are normally used for quantification, since all absorb around 260 nm.
Furthermore, the absorption can be used to detect sample hybridization in solution as a function of temperature, for example. In ssDNA, the bases stack on top of each other, but this conformation is maximized in dsDNA. The hybridization stabilizes the helical structure of DNA and also RNA. Such a feature has been proven to be useful in understanding of the base-pairing of double-stranded DNA [2]. Such a property was used to understand the melting curve of the sequences of DNA-based biosensor development. Figure 2 demonstrates a schematic of a gold electrode surface after the modification with thiolated single-stranded DNA probe with a spacer molecule (6-mercapto-1-hexanol) to control the probe surface density.
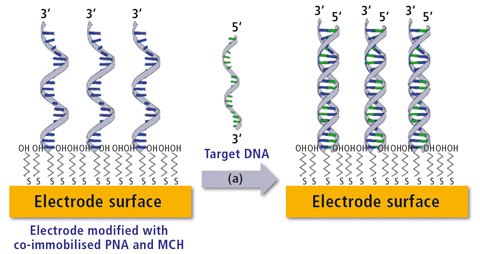 Figure 2: Schematic of a DNA biosensor. Left shows the co-immobilization of thiolated DNA probe with a spacer molecule (6-mercapto-1-hexanol). Right represents post capture of target DNA resulting in the formation of a duplex strand.
Figure 2: Schematic of a DNA biosensor. Left shows the co-immobilization of thiolated DNA probe with a spacer molecule (6-mercapto-1-hexanol). Right represents post capture of target DNA resulting in the formation of a duplex strand.
Methods
For characterization of the nature of hybridization of the probe DNA and target DNA, an 8-series micro cell was used with silicone plugs, which fits inside the TMSPC-8 temperature controlled accessory. A Shimadzu UV-1800 spectrophotometer equipped with the TMSPC-8 was applied for the thermal melt experiment.
1 µM concentration of probe DNA (5’- SH(CH6) -TTTTTATTGTGACAGACCATTGCTACA-3’), target DNA (5’- TGTAGCAATGGTCTGTCACAAT-3’) and the mixture of probe and target DNA was prepared separately in 10 mM phosphate buffer saline (PBS, pH 7.4). The first cell was just used to measure the blank with PBS buffer, followed by the use of two cells each for probe DNA, target DNA and mixture (probe + target) respectively. The program was set with a complete cycle where the temperature was raised from 19 °C to 90 °C and back. Using a ramp rate of 0.5 °C/min, the absorbance was recorded at a wavelength of 260 nm.
Figure 3 shows the schematic of gold nanoparticle (AuNP) before and after modification with polyethylene glycol (PEG). Synthesis of 17 nm AuNP was adopted by the protocol reported by Gao, Jie, et al [3]. Briefly, 5 mL HAuCl4 gold (III) chloride hydrate (0.2 % w/w) from Sigma-Aldrich was added to 90 mL water and was heated to 80 °C with constant stirring. Later, 5 ml of trisodium citrate dihydrate solution (1 % w/w) from Sigma-Aldrich in water was added quickly leading to changing of the color to deep red.
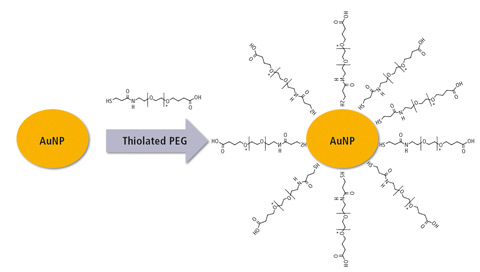 Figure 3: Synthesis of gold nanoparticles (AuNP) followed by modification with polyethylene glycol (PEG)
Figure 3: Synthesis of gold nanoparticles (AuNP) followed by modification with polyethylene glycol (PEG)
Thereafter, the synthesized AuNP was modified by polyethylene glycol (PEG). 1 mL of AuNP was incubated with 100 uL of 0.5 mM (O-(3-Carboxypropyl)-O’-[2-(3mercaptopropionylamino) ethyl] -polyethylene glycol (PEG) from Sigma-Aldrich [MW 3000 Da]) for 16 hours at 4 °C with constant stirring. The modification of AuNP before and after the modification was monitored using Shimadzu’s UV-1800, looking at the absorbance spectrum in the wavelength from 400 to 800 nm.
Results and Discussion
Figure 4 shows the real-time melting curve obtained for the DNA probe and target DNA from 19 °C to 90 °C and back. Two DNA strands hybridize at room temperature and with the increase of temperature, the conformation changes until 60 °C. Up to this value, the double helix denatures, increasing the absorbance. When the temperature decreases, the single strands hybridize again and the absorbance reduces. The temperature at which half of DNA strands are in the single-stranded (ssDNA) state is termed as melting temperature (Tm). From the software analysis, the Tm in 10 mM PBS (pH 7.4) was found to be 66 °C.
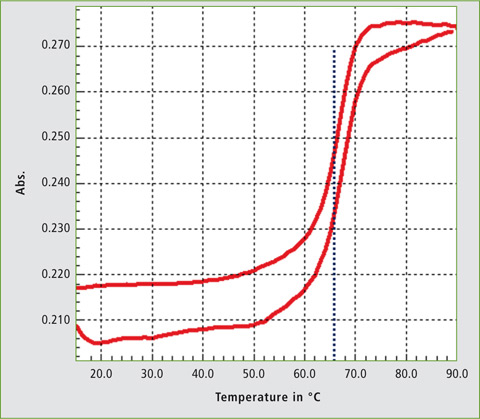 Figure 4: Melting curve of DNA probe and target DNA in 10 mM PBS buffer, pH 7.4
Figure 4: Melting curve of DNA probe and target DNA in 10 mM PBS buffer, pH 7.4
The analysis confirmed that the two strands obtained hybridized well due to their complementary properties. Different nucleotide sequences lead to different denaturation temperatures which can be analysed with UV light. Similarly, hybridization events can be confirmed using this technique.
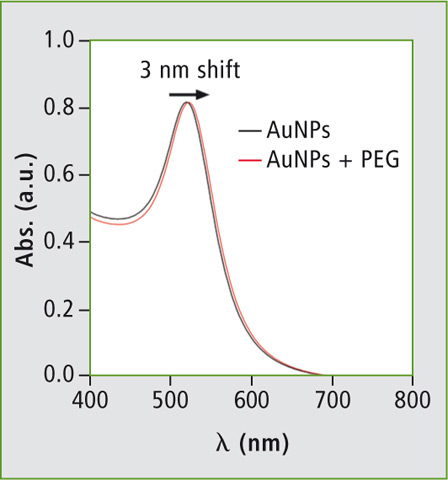 Figure 5: Absorbance curve of gold nanoparticle (AuNP) before and after modification with polyethylene glycol (PEG)
Figure 5: Absorbance curve of gold nanoparticle (AuNP) before and after modification with polyethylene glycol (PEG)
Figure 5 shows the absorbance curve obtained for the synthesised AuNP. The graph shows characteristic peaks of 20 nm AuNPs at 520 nm. After the modification of AuNP with PEG molecules, the characteristic peak shows a 3 nm displacement which is well in accordance with the reported literature [3].
Conclusion
In conclusion, the impetus of this application news was to demonstrate how the Shimadzu UV-1800 together with Thermal Melt Analysis (TMSPC-8) system can be utilized as a characterizing tool to develop a biosensor. The UV-1800, when incorporated with the TMSPC-8 System is capable of monitoring the changes in absorbance of the dsDNA melting as a function of temperature and determining Tm values for the dsDNA in the buffer applied. It was also used to understand the nature of hybridization and to confirm the use of right sequences for the development of DNA-based sensor. UV-1800 was also applied to monitor the changes in absorbance on the modification of gold nanoparticles with PEG. With the 8-channel micro cell, multiple curves can be captured simultaneously for the analysis of repetitive sample. Another advantage of using such a cell is the need of low sample volume (100 µL) enabling analysis of samples in a most efficient way by reducing necessary resources and cost.
Instrumentation
Shimadzu UV-1800 including computer control software UVProbe, high precision cell holder: The TM analysis system (TMSPC-8) is an accessory for UV-Vis, comprising of a thermoelectrically temperature controlled micro multi-cell holder employing a Peltier element and PC software specially designed for the purpose.
Authors
Pawan Jolly 1, Marina Batistuti 2, Pavel Zhurauski 1, Pedro Estrela 1
1 University of Bath, UK; 2 University of São Paulo, Brazil
Literature
[1] Bhalla N, Jolly P, Formisano N & Estrela P. “Introduction to Biosensors”, Essays in Biochemistry. Volume 60 (1), p. 1. 2016
[2] Boyer, Rodney F. “Modern Experimental Biochemistry.” Second Edition. The Benjamin/Cummings Publishing Company. 1993
[3] Gao, Jie, et al. “Colloidal stability of gold nanoparticles modified with thiol compounds: bioconjugation and application in cancer cell imaging.” Langmuir. Volume 28 (9), p. 4464. 2012