New approach for epitranscriptomics
New approach for epitranscriptomics
Quantifying RNA methylations by LC-MS/MS
Dr Alexandre David, Amandine Amalric, Aurore Attina, Pr Sylvain Lehmann, Dr Jerome Vialaret, Pr Christophe Hirtz, Clinical Proteomics Platform, IRMB-PPC, INM, Univ Montpellier, CHU Montpellier, INSERM CNRS, Montpellier, France






Clinical Proteomics Platform, IRMB-PPC, INM, Univ Montpellier, CHU Montpellier, INSERM CNRS, Montpellier, France
Epitranscriptomics is an emerging and promising research field covering biochemical RNA modifications within a cell. It comprises more than 170 RNA chemical marks, including RNA methylation which can regulate the expression of defined genes, and it is present in all type of RNA, such as messenger RNA (mRNA).
Today, mass spectrometry coupled to a liquid chromatography (LC-MS/MS) has become a method of choice to comprehensively measure modified nucleosides from a biological sample. This application note presents a methodology for quantifying methylated nucleosides from cellular mRNA using LC-MS/MS (Shimadzu LCMS-8060).
Introduction
Epitranscriptomics investigates modifications to RNA, which is involved in the production of proteins. This branch of research makes it easier to understand the connection between RNA modifications and diseases. For example, the epitranscriptome is modified in many tumors when compared to healthy cells.
Why is epitranscriptomics so promising?
- There is a high number of RNA modifications with more than 170 RNA chemical marks described so far, including RNA methylation
- These modifications are present in all RNA species (mRNA, tRNA, rRNA, ncRNA and so on)
- RNA modifications are involved in all steps of post-transcriptional regulation of gene expression: RNA structure, splicing, export; stability, cellular localization and translation
- they are involved in key cellular functions such as survival, growth and differentiation.
The N6-methyladenosine nucleoside (m6A) – the most common and abundant modifications on RNA in eukaryotes (i.e. organisms whose cells have a nucleus) – is a striking example. This modification occurs not only on mRNA but also on noncoding RNA (rRNA, tRNA, snRNA). As m6A is important for mRNA regulation in normal physiological condition, any dysregulation of its level can lead to disease. [1]
Current technologies to detect m6A modification [2] use next-generation sequencing techniques and deliver highly valuable information in term of m6A mapping at the genome-wide level. However, they cannot provide precise quantitative results. Today, mass spectrometry coupled to a liquid chromatography (LC-MS/MS) has become the method of choice for accurately quantifying modified nucleosides from biological samples. This application note presents a method for precise quantification of methylated nucleosides from cellular mRNA using LC-MS/MS (Shimadzu LCMS-8060).
| Molecules | Transitions | Retention time (min) | Target Dwell Time (msec) | Target Collision Energy (E) | Target Q1 Pre Bias (V) | Target Q3 Pre Bias (V) | Interface Voltage (kV) |
|
|
268.0 > 136.0 | 11.8 | 197.0 | -18.0 | -24.0 | -24.0 | 1.0 |
| 2’O-methyladenosine (Am) | 282.0 > 136.0 | 14.6 | 397.0 | -17.0 | -14.0 | -24.0 | 1.5 |
| N1-methyladenosine (m1A) | 282.1 > 150.1 | 5.4 | 197.0 | -21.0 | -12.0 | -26.0 | 1.5 |
| N6-methyladenosine (m6A) | 282.1 > 150.1 | 15.9 | 397.0 | -20.0 | -12.0 | -16.0 | 1.5 |
| N6,N6-dimethyladenosine (m6,6A) | 296.0 > 164.1 | 18.3 | 530.0 | -25.0 | -22.0 | -20.0 | 1.5 |
| N6,2’O-dimethyladenosine (m6Am) | 296.0 > 150.0 | 17.6 | 397.0 | -15.0 | -18.0 | -32.0 | 1.5 |
|
|
244.1 > 112.0 | 3.8 | 197.0 | -12.0 | -17.0 | -29.0 | 1.0 |
| 2’O-Methycytidine (Cm) | 258.1 > 112.0 | 7.9 | 142.0 | -15.0 | -18.0 | -32.0 | 1.5 |
| N3-methylcytidine (m3C) | 258.1 > 126.0 | 4.5 | 197.0 | -13.0 | -10.0 | -14.0 | 1.5 |
| N5-methylcytidine (m5C) | 258.0 > 126.0 | 7.3 | 157.0 | -17.0 | -14.0 | -25.0 | 1.0 |
|
|
284.1 > 152.0 | 8.6 | 142.0 | -15.0 | -24.0 | -23.0 | 1.5 |
| 2’O-methylguanosine (Gm) | 298.1 > 152.0 | 10.9 | 174.0 | -12.0 | -12.0 | -17.0 | 1.5 |
| N1-methylguanosine (m1G) | 298.1 > 166.0 | 10.6 | 142.0 | -15.0 | -11.0 | -18.0 | 0.5 |
| N7-ethylguanosine (m7G) | 298.3 > 166.0 | 7.3 | 157.0 | -10.0 | -30.0 | -18.0 | 1.5 |
| N2,N7-dimethylguanosine (m2,7G) | 312.1 > 180.0 | 10.3 | 142.0 | -12.0 | -12.0 | -17.0 | 2.5 |
| N2,N2,N7-trimethylguanosine (m2,2,7G) | 326.15 > 194 | 12.4 | 197.0 | -20.0 | -15.0 | -21.0 | 1.5 |
|
|
245.1 > 113.0 | 5.2 | 197.0 | -11.0 | -28.0 | -26.0 | 1.0 |
| 2’O-methyluridine (Um) | 259.1 > 113.0 | 9.5 | 142.0 | -9.0 | -11.0 | -19.0 | 2.5 |
Pre-analytical steps
Experimental workflow for quantification of methylated nucleosides by LC-MS/MS is shown in figure 1.
106 cells CTC44 (Circulating Tumor Cells) were seeded into 150 mm x 15 mm petri dishes and cultured for 4 days at 37 °C under humidified 5 % CO2 in high glucose DMEM (Dulbecco’s Modified Eagle’s Medium) supplemented with 2 mM glutamine and 10 % FCS (Fetal Calf Serum).
RNA extraction was achieved using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. mRNA was isolated twice using an GenElute mRNA purification kit (Sigma). First, 400 ng of RNA (in 20 µL final) was digested by 5 units of RNA 5’ pyrophosphohydrolase (New England Biolabs) for 2 h at 37 °C with NEB buffer. Then decapped RNA was digested into nucleotides by 1 unit of nuclease P1 (Penicillium citrinum, Sigma) for 2 h at 42 °C on NH4OAc buffer (10 mM, pH 5.3). Finally, nucleotides were dephosphorylated into nucleosides using 1 unit of alkaline phosphatase (Escherichia coli, Sigma) for 2 h at 37 °C on NH4OAc buffer (100 mM). [3]
After adding 60 µL of phase A (ammonium acetate, pH 5.3), the samples are filtered (0.22 μm pore size, 4 mm diameter, Millipore), and 5 μL of the solution was injected into LC-MS/MS.
Analytical conditions
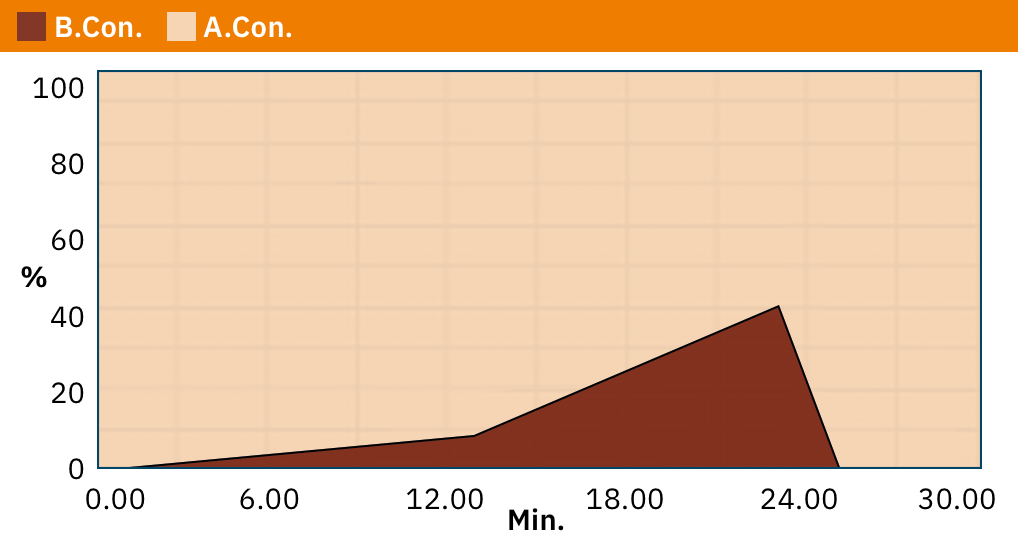
For the measurement of nucleosides using LC-MS/MS, the nucleosides are separated by reverse phase UHPLC (ultra-performance liquid chromatography) and detected by the Shimadzu LCMS-8060 triple-quadrupole mass spectrometer in multiple reactions monitoring (MRM) positive electrospray ionization (ESI) mode. Details of analysis conditions and MRM parameters are given in tables 1 and 2.
Modomics Database [4] contains information of MRM transitions of numerous modified nucleosides, and enables simultaneous measurement of several methylated nucleosides.
Quantification of methylated nucleosides from messenger RNA (mRNA)
The quantification of methylated nucleosides from mRNA yielded results for ten nucleoside species from the target panel of 14 RNA methylations (figure 3). The remaining four species have only been observed in rRNA, tRNA, and snRNA, and are – to the author’s knowledge – not present in mRNA (figure 2).
Furthermore, for most of the methylations detected, a lower signal is observed for mRNA compared to total RNA (figure 4). For example, m1A is found in mRNA, which accounts for only 5 % of the total cell RNA, and also in tRNA and rRNA, which respectively represent 15 % and 80 %.
In the case of mRNA purification, the ion signal was enhanced for methylations that are mostly predominantly found in mRNA, such as m6A and m6Am (figures 4 and 5).
Analytical conditions
For the measurement of nucleosides using LC-MS/MS, the nucleosides are separated by reverse phase UHPLC (ultra-performance liquid chromatography) and detected by the Shimadzu LCMS-8060 triple-quadrupole mass spectrometer in multiple reactions monitoring (MRM) positive electrospray ionization (ESI) mode. Details of analysis conditions and MRM parameters are given in tables 1 and 2.
Modomics Database [4] contains information of MRM transitions of numerous modified nucleosides, and enables simultaneous measurement of several methylated nucleosides.
Quantification of methylated nucleosides from messenger RNA (mRNA)
The quantification of methylated nucleosides from mRNA yielded results for ten nucleoside species from the target panel of 14 RNA methylations (figure 3). The remaining four species have only been observed in rRNA, tRNA, and snRNA, and are – to the author’s knowledge – not present in mRNA (figure 2).
Furthermore, for most of the methylations detected, a lower signal is observed for mRNA compared to total RNA (figure 4). For example, m1A is found in mRNA, which accounts for only 5 % of the total cell RNA, and also in tRNA and rRNA, which respectively represent 15 % and 80 %.
In the case of mRNA purification, the ion signal was enhanced for methylations that are mostly predominantly found in mRNA, such as m6A and m6Am (figures 4 and 5).
Conclusion
RNA methylations label free quantification using a triple quadrupole LC-MS/MS system (LCMS-8060 mass spectrometer coupled to Nexera LC-40 in MRM mode) represents a sensitive and reliable approach for epitranscriptomics studies. This application can be implemented for a large panel of biological samples (and RNA species). It is a straightforward and readily applicable experimental setup.
Literature
- C. Yang et al. The role of m6A modification in physiology and disease, Cell Death Dis (2020). doi: 10.1038/s41419-020-03143-z
- X. Li et al. Epitranscriptome sequencing technologies: decoding RNA modifications, Nat Methods (2016). doi: 10.1038/nmeth.4110
- S. Relier et al. FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat Commun 12, 1716 (2021). https://doi.org/10.1038/s41467-021-21758-4
- genesilico.pl/modomics
- https://onlinelibrary.wiley.com/doi/full/10.1002/nadc.20174060797
- https://www.mpg.de/12621941/psych_jb_2018
- https://www.lungeninformationsdienst.de/forschung/epigenetik/forschung/index.html
- https://www.tamaserv.com/angebot